A
TUTORIAL ON HOW TO USE BIOMART IN ENSEMBL
BIOMART :
In this video I have explained the usage of
BIOMART. It is a simplest online tool that permits extraction of information
with no programming information or comprehension of the basic database
structure.
HOW
TO USE A BIOMART:
1.
Selection of Database Type
Following are the
databases that can be chosen :
- Ensembl gene
- Ensembl variation
- Ensembl regulation
- Vega
2.
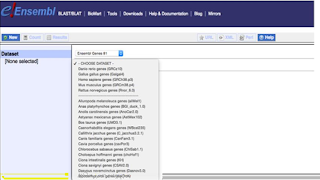
Selection of Database Species
Data
available for all the Species including Ensembl genes,Ensembl variation,
Ensembl regulation and Vega.
3.
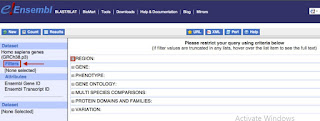
Filtration of Input Data
- BioMart permits you to confine your question with data that you know, e.g: input a list of IDs, restrict to a region.
- You can access the filter page by clicking on the "Filters" button located on the left panel.
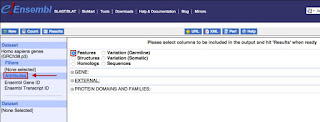
4.Selection
of Attributes
This option permits you
to choose your ideal yield; the default yield is
- Ensembl Gene ID
- Ensembl Transcript ID
Thank you
Waiting for your valuable response

Comments
Post a Comment